A quick update on what I'll be working on over the next few days.
James, Dr. Shaun, Ada and I had a call regarding PTM 47, which is aimed at moving the patientmatching module reports from a flat file system to the database.
This is going to be a complex task, but I feel that our call ironed out a lot of issues / complications that we were facing.
We are going to move the files into the database in two major stages. This will be keeping me very busy over the next few weeks.
More details on the ticket can be found here.
Monday, December 19, 2011
Sunday, December 18, 2011
I've been published in the Sunday Observer !!
Some more (slightly delayed) good news.
A article on myself and my OpenMRS work was published in this weeks Sunday Observer.
Now for those of you who don't know, the Sunday observer is Sri Lankas leading State owned English newspaper.
As I remember, their circulation is about 0.5 million copies (which is a very big deal in a country such as Sri Lanka.)
Basically, the article covers my comments on my OpenMRS work, and also some tips / hints on working in the community.
In fact, its the very same article on myself that was published in the IIT Student magazine. I cant remember if I ever blogged on that article, but let me check up on that.
I'm still looking for an online version of this item, but failing to find one, Ill upload a photo of the article.
But anyway, this article in not an actual 'victory', but rather a report of an old victory. Lets hope there'll be lots more victories over the next year ! :-)
PS: On an unrelated note, I just remembered that one of my key concerns during my days as a writer was how much longer it would take for an article about myself (as opposed to an article written by myself) to get published in a newspaper. Hmm...... So my pen name first appeared on a national newspaper at the age of 19. The first article about me was published when I'm 24. So, 24 -19 = 5 years :-)
A article on myself and my OpenMRS work was published in this weeks Sunday Observer.
Now for those of you who don't know, the Sunday observer is Sri Lankas leading State owned English newspaper.
As I remember, their circulation is about 0.5 million copies (which is a very big deal in a country such as Sri Lanka.)
Basically, the article covers my comments on my OpenMRS work, and also some tips / hints on working in the community.
In fact, its the very same article on myself that was published in the IIT Student magazine. I cant remember if I ever blogged on that article, but let me check up on that.
I'm still looking for an online version of this item, but failing to find one, Ill upload a photo of the article.
But anyway, this article in not an actual 'victory', but rather a report of an old victory. Lets hope there'll be lots more victories over the next year ! :-)
PS: On an unrelated note, I just remembered that one of my key concerns during my days as a writer was how much longer it would take for an article about myself (as opposed to an article written by myself) to get published in a newspaper. Hmm...... So my pen name first appeared on a national newspaper at the age of 19. The first article about me was published when I'm 24. So, 24 -19 = 5 years :-)
Sunday, December 11, 2011
Mutual Scoring enhancements are committed !!
So, finally, after months of sweat and toil, Mutual Scoring (or MI for short) was committed today.
To me, this represents a major milestone in my internship. Firstly, it marks my involvement in real research work with the top minds in the industry. (to put it more clearly, I can now say that I've "been there, done that, and got the tee shirt"). Secondly, it marks my commitment to this industry, and what it has to offer, and finally, that i'm doing my best to follow the advice given by Professor Szolovits, and am following in at least the general direction that he suggested.
Of course, I admit that there is a lot more that I can do, but I still have two more years until I graduate, so hopefully I'll get better.
You're welcome to check out the changes in the patientMatching module. Right now, these enhancements are very 'nerd' friendly, but not very user friendly.
I'm planning to take care of that along with the second phase of mutual scoring, which i'm hoping to start work on as soon as I take care of a few course works. (mustn't forget that I have to finish my degree as well :-)
So right now, I can happily say that phase one of MI scoring is over, and I can return to the drawing board for phase two.
Im indebted to the support given by Dr. Grannis (THE Dr. Shaun Grannis) and James Egg of Regenstrief.. I could never have done this without them.
Best Regards,
Suranga
To me, this represents a major milestone in my internship. Firstly, it marks my involvement in real research work with the top minds in the industry. (to put it more clearly, I can now say that I've "been there, done that, and got the tee shirt"). Secondly, it marks my commitment to this industry, and what it has to offer, and finally, that i'm doing my best to follow the advice given by Professor Szolovits, and am following in at least the general direction that he suggested.
Of course, I admit that there is a lot more that I can do, but I still have two more years until I graduate, so hopefully I'll get better.
You're welcome to check out the changes in the patientMatching module. Right now, these enhancements are very 'nerd' friendly, but not very user friendly.
I'm planning to take care of that along with the second phase of mutual scoring, which i'm hoping to start work on as soon as I take care of a few course works. (mustn't forget that I have to finish my degree as well :-)
So right now, I can happily say that phase one of MI scoring is over, and I can return to the drawing board for phase two.
Im indebted to the support given by Dr. Grannis (THE Dr. Shaun Grannis) and James Egg of Regenstrief.. I could never have done this without them.
Best Regards,
Suranga
Monday, December 5, 2011
My tee shirts have arrived !
So, as the heading says, my tee shirts arrived today. As anticipated, the Sri Lankan postal department had torn open the parcel to see if they can steal anything. Unfortunately for them, the parcel contained only items of sentimental value (I had especially asked Ada NOT to send anything valuable) So all the contents of the parcel were pretty much intact.
I got the Muzungu shirt (mentioned in previous post) an AMPATH tee shirt and finally, an awesome Regenstrief tee shirt.
The tee shirt is rather big, but I really really love it.
I'll post a pic of me wearing this shirt here. Please excuse my unusual apperance in this photo. It was taken shortly after I just work up :-)
Thanks to everyone who helped get the shirts from Kenya -> Indiana -> Sri Lanka !!!!!!!!!!
Tuesday, November 15, 2011
Hectic Times
I've been unable to update this blog regularly because, as you can see by my title, I've been going through hectic times.
Firstly, i'm still very much involved in AMPATH work - so much so that I've been unable to spend any time with my beloved OpenMRS core over the past month or so.
However, We've definitely making progress. Ada is even getting me an AMPATH tee shirt (So that I can say 'I've been there, and done that'. ) Poor Ben (our Director) has been asked to do the purchasing (since he's frequently flying to and fro between Eldoret and Indy).
I'm also getting an Muzungu tee shirt. Muzungu is basically African for 'the white people' (in Swahili is means 'confused person wondering about' :-D ).
I guess Muzungu Tee shirts are somewhat of a fad of Regenstrief and PIH employees. I think its also very popular among westerners working in Africa. But anyway, I'm really looking forward to the tee shirts, and hoping that Ben wont get them in the lemon green and pink colours that he suggested :-)
Other than this, I'm also working on two other OpenMRS related projects, as part of something that Professor Solovitz of MIT suggested. Dr. Mamlin is helping me with one of these, while my favorite lecturer from IIT is helping with the other. (well, formerly IIT, and now with the Swinburne University, AUS).
Hopefully, we'll know the outcome of these efforts by Dec - Jan. But until that, its all confidential.
I'm also trying to continue with Dr. Shauns' Patient matching improvements, but its been really hard to find the free time.
And plus, my university does not understand that I'm really busy, and keeps assigning Course works.
Oh well.......
Labels:
AMPATH,
Ben,
modules,
Muzungu,
OpenMRS,
PIH,
Regenstrief,
Shaun Grannis
Friday, November 11, 2011
My work with AMPATH - Part two
It's been some time since I published part 1 of my AMPATH story. I couldn't follow up with part two because the project is still very much ongoing, and I've only just committed the latest changes....
But let me try to pick up this story where I left off, and write at least a chapter or so on our progress.
Well, (resuming our narrative...) Ada tried out my new omod on an openmrs server, and sure enough it worked.
So she went ahead and uploaded the new module to the AMPATH research server, and suddenly it did not work.
The error was as follows,
Error Starting Module – Patient Matching
Error while trying to start module
Error while running sql: DROP TABLE IF EXISTS `openmrs`.`persistreportdata` . Message: DROP command denied to user 'amrs_user'@'localhost' for table 'persistreportdata'
I has just started digging into this problem when I got a mail from Win (Nyoman Ribeka) a Regenstrief developer and one of the fist contributors (well, actually THE first contributor) to the patientmatching module.
Apparently Ada had forwarded him the stack trace, and it had taken Win all of five minutes to come up with the solution.
The problem was that we had tested out the fixes using an 'openmrs' database, while the AMPATH database was named 'amrs' ! . The issue was being caused by a database dependent code snippet that had somehow by-passed our vigorous code review process.
So naturally our fixes worked on the openmrs test environment, and crashed on the AMPATH environment.
But anyway, it was quite easy to fix once we understood the cause. we traced the faulty code to the sqldiff file ( an SQL inset into statement had the database name 'openmrs' prepended to the end. This was unnecessary, and was causing our problems. )
So I fixed it, tested it out and sent Ada the latest omod...
And was that the end of our problems ? not quite,
But that is for later. Do stay tuned for part -3 !
Thursday, November 3, 2011
My work with AMPATH - Part one
I've been busy working with AMPATH engineers over the past week. We did a lot of important work to help AMPATH switch to the latest patient Matching module, and I'm happy to have played a role in getting this up and running.
For all of you who don't know what AMPATH is, its practically one of OpenMRS's birthplaces. For more details, please visit here.
It all started off after I got a mail from Dr. Shaun (THE Dr. Shaun from Regenstrief) asking if I could help the AMPATH people upgrade their system to the latest patient matching release. basically, AMPATH was using OpenMRS 1.8.2, and wanted to move on to the latest version of our module (which was supposed to be 1.2.2, but ultimately turned out to be 1.2.4 due to all the changes that we made)
Naturally, I was pleased to jump in and help out (who wouldn't ?)
So I contacted Ada (Ada Yeung, who works with AMPATH) got access to AMPATH test server, and started checking out the system. Unfortunately, the test server was somewhat muddled up, mainly due to the large number of tomcat and OpenMRS modules installed everywhere.
So I contacted James (James Egg from Regenstrief) sorted everything out, upgraded the test server and got the latest running. The Upgrading process went pretty smoothly ( You will really admire the OpenMRS design once you see how easy it is to switch to - fro and upgrade with a live DB) and I didnt have any data problems at all.
So I went ahead and fixed a number of major showstoppers.
Dr. Shaun and Ada were very happy with my work, especially with the bug fixes.
So I committed the new changes, and stood down for the testing to begin.
And so was that the end of the story ? oh no, this was just the beginning. It all got pretty complicated after that...
Expect the full details in the next blog post !!!!
Thursday, October 27, 2011
I've just been published by Google
A quick post to say that I've been published on the Google blog.
More specifically, an article I wrote on my experience at the OpenMRS implementer conference was published on the Google opensource blog.
This is a really big deal to me, since its the first time I have been singly picked out by Google. However a lot of the credit should go to my good friend Michael Downey, who actually spearhearded the entire effort. If it wasn't for him, I'd be just another boring old student.
The article appears here (http://google-opensource.blogspot.com/2011/10/openmrs-welcomes-with-open-arms.html)
Its been modified slightly so that none technical people can understand it better, but otherwise its pretty much the same.
PS: I can honestly say that this is the "one small step for me, one huge step for my future" moment of my career.
Thank you Michael !!!!!
More specifically, an article I wrote on my experience at the OpenMRS implementer conference was published on the Google opensource blog.
This is a really big deal to me, since its the first time I have been singly picked out by Google. However a lot of the credit should go to my good friend Michael Downey, who actually spearhearded the entire effort. If it wasn't for him, I'd be just another boring old student.
The article appears here (http://google-opensource.blogspot.com/2011/10/openmrs-welcomes-with-open-arms.html)
Its been modified slightly so that none technical people can understand it better, but otherwise its pretty much the same.
PS: I can honestly say that this is the "one small step for me, one huge step for my future" moment of my career.
Thank you Michael !!!!!
Labels:
conference,
google,
Kigali,
meeting,
Michael Downey,
OpenMRS
Thursday, October 20, 2011
OpenMRS patient Matching module
I've recently started work on the OpenMRS patient Matching module. Its a complex and intriguing piece of software planned by Dr. Shaun Grannis of the Regenstrief institute.
Dr. Shaun is qualified in both engineering as well as medicine (he's an MIT graduate) and his mathematical training came really helpful in my efforts. I'm also being helped by James Egg, also of the Regenstrief institute. He has been a great help with design planning and domain expertise.
I also found out that Win (Nyoman Ribeka) of OpenMRS had initially got involved in OpenMRS working on the PatientMatching module as a gsoc student ! (this was in 2008)
My work on the module is aimed at calculating mutual information scores for pairwise combinations of columns. This will ultimately make the matching algorithm more efficient.
The work is quite challenging, but intriguing and interesting. I have already made much progress in my work.
for further reference, documentation on the module is available on the wiki here
Credit must also go to Gautami, a fellow Gsoc student who worked on this module during Gsoc 2011.
Dr. Shaun is qualified in both engineering as well as medicine (he's an MIT graduate) and his mathematical training came really helpful in my efforts. I'm also being helped by James Egg, also of the Regenstrief institute. He has been a great help with design planning and domain expertise.
I also found out that Win (Nyoman Ribeka) of OpenMRS had initially got involved in OpenMRS working on the PatientMatching module as a gsoc student ! (this was in 2008)
My work on the module is aimed at calculating mutual information scores for pairwise combinations of columns. This will ultimately make the matching algorithm more efficient.
The work is quite challenging, but intriguing and interesting. I have already made much progress in my work.
for further reference, documentation on the module is available on the wiki here
Credit must also go to Gautami, a fellow Gsoc student who worked on this module during Gsoc 2011.
Meeting the OpenMRS community at Kigali, Rwanda
I've been unable to post much on my blog recently. The reason for this was that I was away at Kigali, Rwanda, attending the annual OpenMRS implementer conference.
So much things happened there that I've been tied up with a lot of important //TO DOs that kept me busy for a while.
but first and foremost, I'd like to share an article I wrote on the event. This was posted on the OpenMRS blog here.
However i'm re posting it here because I really like it. Enjoy !

So much things happened there that I've been tied up with a lot of important //TO DOs that kept me busy for a while.
but first and foremost, I'd like to share an article I wrote on the event. This was posted on the OpenMRS blog here.
However i'm re posting it here because I really like it. Enjoy !
Google Summer of Code Student Meets the OpenMRS Community
Recently, I had the rare privilege of attending the annual implementers conference of OpenMRS, my Google Summer of Code (GSoC) mentoring organization.
Thanks largely to the conference sponsorship by Google, OpenMRS was able to fund my week-long visit to Kigali, Rwanda. I travelled over 4,500 miles to Rwanda, and spent a week meeting with core OpenMRS developers, other volunteers, implementers, service providers and researchers who had gathered there for the conference.

OpenMRS Implementers Meeting 2011
Arriving at Kigali, I was amazed at the diversity and fellowship amongst the community, and of how happy they were to accept me as their own.
I took part in a pre-conference hackathon, and visited the OpenMRS implementation at Rwanda’s TRAC Plus health clinic. I listened to why implementers from Village Health Works used an Access database for their Burundi clinic, and to Dr. Joaquin Blaya’s work with Interactive Voice Response. Eduardo Jezierski, the CTO of InSTEDD, talked to me about their work in Haiti, while Christopher Bailey of the World Health Organization spoke of his experience working with developing countries.
I also enjoyed a Chinese meal with a group of US-based developers for the AMPATH program in Western Kenya. (Ben Wolfe, director of OpenMRS API Development taught me how to use chopsticks). I visited the Kigali Genocide museum, played cards with a group of research scientists, academics and other developers and spilled my drink all over a director’s laptop. I had dinner with OpenMRS co-founder Dr. Burke Mamlin, brought a drink for co-founder Dr. Paul Biondrich and had breakfast with an MIT graduate who explained why he quit building space satellites and an engineering career to enroll in medical school.
My participation at the conference was an eye opener for several reasons. Firstly, it helped me make the change from “GSOC student” to fully fledged community member. I stopped being just an offshore volunteer, and understood my organization for what it really was.
I saw my project as a community, a group of vibrant, talented and extremely capable people with a wide range of interest in software development, research, medicine, health informatics and public health. I understood that OpenMRS is not “just” OpenMRS, but a massive network of implementers, developers, healthcare workers and other organizations.
I saw the dedication and professionalism of community members, and their sincere concern to help make the world a better place. I also realized how community members were supporting themselves while enjoying what they do by serving as consultants, developers and service providers.
However, I believe that my experience at the conference highlights something that Google should definitely look into – the inability to ensure that GSoC students continue being a part of the open source community once deadlines are over. As Google clearly states on their website, GSoC is organized with the aim of encouraging students to contribute to open source projects. However it’s undeniable that many students consider GSoC as a get rich scheme for their internship year.
I feel that this is mainly due to four reasons – firstly because many GSoC students are offshore based, and see their project as yet another software, secondly because many of them consider it as a internship with strict deadlines, thirdly because they are unable to make the transition from volunteer to community member and finally because they fail to understand that open source developers have ways of making money while doing what they enjoy.
As I see, the best way that Google can avoid this is by tearing down the wall between student and community member.
Google can do this by introducing more flexible methods of student evaluation, by giving more weight to community participation in their evaluation criteria and possibly by moving up student application dates so that students can apply several months in advance, allowing the community to assess how they get up to speed on domain knowledge and technology stacks. This way, students will be familiar with their projects, the community will be aware of their capabilities and stronger ties would link the student with his organization.
Ideally, all this would happen before the student is even selected for GSoC.
Ultimately, I believe that both Google and mentoring organizations should consider methods to ensure that GSoC does not end with “successful project completion”. Also consider – why do we even get a certificate saying that we “are recognized for participating in GSoC”? It should say that we are “recognized as a successful community member” of our respective mentoring organizations.
Saturday, October 1, 2011
OpenMRS Sync module
I've been involved in the OpenMRS sync module.
We had a sprint for this a few weeks back. Basically, we were aiming to prepare it for a 1.0 release.
The sync module is great concept. You can check it out here.
Its purpose is to let users who have multiple sites with separate databases to copy data from each other. Databases are defined as parent or child databases.
The Parent is always the computer or site that is most central. Multiple child sites send their data up to the parent site for dissemination. Children/parent sites can be set up in a multi hierarchy way.
Apparently work on the Sync module was started as far back as 2007 by Christian Allen of PIH.
Its also noteworthy that the Sync module is the first ever OpenMRS module that i'd been involved in - all my prior work was on the trunk.
Even more noteworthy, not a single one of my commits (thankfully) broke the build during the sprint .....
Labels:
modules,
OpenMRS,
sprint,
sync module
Thursday, September 29, 2011
OpenMRS implementers meeting 2011
I'm pleased to announce the OpenMRS implementers meeting of 2011.
This is an annual conference where OpenMRS contributors and implemnters meet up to share ideas, make plans and to improve ourselves and our community.
This years conference will be held in Kigali, the capital city of Rwanda. I am able to attend because the OpenMRS project has kindly come forward to sponsor my visit. They will be taking care of my finances.
I am really looking forward to the meeting, because I will get to meet a lot of the best people in the industry. this is sure to be a great learning experience for me. Hopefully, I will also take part in the hackathon held prior to the event.
It'll be great to meet the Dev group, Ben, Darius, my dear friend Daniel, Michael and Rafa. I will also meet up with some other people from Ampath, PIH and Eldoret.
Dr. Mamlin and Dr. Biondritch will also attend. So will Dr. Hamish Fraser and Dr. Shaun Grannis of the Regenstrief institute.
It'll be great to meet all these people for the first time.
Unfortunately, my mentor Glen McCallum will be a notable absentee at the event, so I wont be seeing him at Kigali.
PS: Not to forget Dr. Joaquin, who has a PhD from HST. He has been one of my major advisers, so I'm looking forward to meeting him as well.
See here for further details on the meeting.
Labels:
2011,
conference,
Kigali,
meeting,
OpenMRS,
Regenstrief
A few of my early ideas
Pasted below are a few of my earliest ideas for health informatics related projects. I'm posting them here for future reference. Hopefully, one day I'll be able to implement much better ideas....
I’ve been studying syndromic surveillance recently.
I studied this [0] document, which addresses shortcomings in syndromic surveillance. It may be rather outdated, but I feel that it raises a strong argument regarding what we’re doing wrong, and what needs to be improved.
Tradeoffs in many surveillance systems are
I’ve also considered,
Is it easier to work with more data, or less data? Can it be useful, or isn’t it worth the effort?
Another major objective in my mind-
You don’t need a system that will take up much resources, and finally say “ok, we calculate that you have an epidemic happening right here, right now”. Instead, we need something that says, “I’m detecting patterns which indicate a 50% chance of epidemic proportions in location A by next Monday. Based on these results, you need to keep an eye on Location B as well, since I’ve detected emerging patterns over there too.”
Syndromic surveillance tools should be able to identify any emerging illness pattern, not just biological attacks.
Ultimately, there is a limit to what an artificial system can do. I believe that the final decision regarding whether or not to issue an epidemic alert should lie with a medical practitioner. The surveillance systems’ purpose it to provide the practitioner with meaningful information that will help him take a decision. Nothing more, nothing less.
Decentralization
What are the tradeoffs between decentralizing and converging of syndromatic surveillance?
What’s the impact of considering data from a wide range of locations if we’re dealing with an epidemic that’s spread across a very small demographical region? Will this help us, or will it cause problems?
In my opinion, centralizing, or converging all health data into a single processing system encourages the possibility of a single point of failure.
On the other hand, de centralizing will improve the system by offering it not one, but several chances to succeed. It will also let us create a tool that will support both pandemic and epidemic surveillance.
Another point – check out this [1] an article on ‘Rapid detection of pandemic influenza in the presence of seasonal influenza’
My question is, how do we identify if it’s just a seasonal ‘spike’ or an emerging epidemic? And how do we map symptoms to specific illnesses, and make that match before it’s too late?
So how to detect an emerging outbreak?
I came up with the following diagram to track patients and their symptoms for hospital A.
The three types of images given here depict different symptoms.
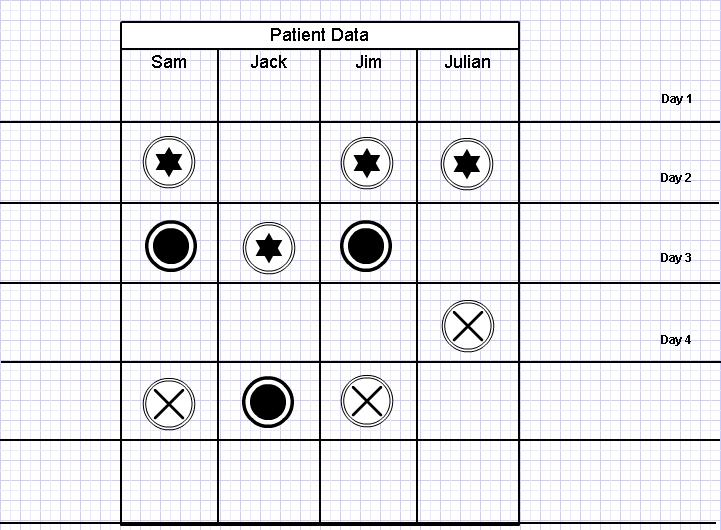
Assume the cases of Sam, Jack, Jim and Julian.
On Day 2, Sam, Jim and Julian all develop symptom A, which is associated with flu. So does this mean that we are facing an influenza epidemic? Maybe, maybe not. But on day three, Sam and Jim both develop symptom B. So now we have a pattern.
We also note that Sam and Jim go on to develop Symptom C on day 5. So this would definitely mean that Sam and Jim are suffering from the same illnesses.
But consider Jack. He developed symptom B a day later than normal. So what are his chances of suffering from the same illness as Sam and Jim?
And also, it seems that Julian has developed symptom A and C, but not B.
So based on this, and assuming that the symptoms are for flu, we could hypothetically come up with the following.
So now we have a pattern, and we can stay alert for it. This pattern can also be used together with mathematical disease modeling to watch for outbreaks at other locations as well. This works well with the centralized (converged) vs. decentralized surveillance model I described earlier.
For example, assuming we have five data sources (hospitals) and one central surveillance unit.
If the unit fails to identify an epidemic incident in source A, it will essentially fail to identify similar cases of epidemics in the remaining four sources as well.
But if de centralized, we are improving the chances of epidemic detection by five times.
Example: assuming that our system will detect an epidemic in source A, but fail to do so in other four sources.
Once the pandemic is identified in source A, the unit will extract a set of core ‘definers’ that will be sent to the other units for further processing. Each of the other centers will do a backwards check to see if there is an indication of such a pattern in symptoms reported to them. Not only do these centers get a second chance to check for mistakes, they also get a pre warning if an outbreak is imminent.
And assuming that a disease has only just infected the region, a different location may have only received reports of symptom A so far. If so, due to the indicators sent from other regions they will be able to make preparations for a possible outbreak depending on the emergence of symptom B as well.
We may even share the core definers with other hospitals to do a wider ranging check.
[0] http://www.rand.org/pubs/research_briefs/RB9042/index1.html
[1] http://www.biomedcentral.com/content/pdf/1471-2458-10-726.pdf
I studied this [0] document, which addresses shortcomings in syndromic surveillance. It may be rather outdated, but I feel that it raises a strong argument regarding what we’re doing wrong, and what needs to be improved.
Tradeoffs in many surveillance systems are
- Sensitivity
- Timeliness
- False positive rates
I’ve also considered,
Is it easier to work with more data, or less data? Can it be useful, or isn’t it worth the effort?
Another major objective in my mind-
You don’t need a system that will take up much resources, and finally say “ok, we calculate that you have an epidemic happening right here, right now”. Instead, we need something that says, “I’m detecting patterns which indicate a 50% chance of epidemic proportions in location A by next Monday. Based on these results, you need to keep an eye on Location B as well, since I’ve detected emerging patterns over there too.”
Syndromic surveillance tools should be able to identify any emerging illness pattern, not just biological attacks.
Ultimately, there is a limit to what an artificial system can do. I believe that the final decision regarding whether or not to issue an epidemic alert should lie with a medical practitioner. The surveillance systems’ purpose it to provide the practitioner with meaningful information that will help him take a decision. Nothing more, nothing less.
Decentralization
What are the tradeoffs between decentralizing and converging of syndromatic surveillance?
What’s the impact of considering data from a wide range of locations if we’re dealing with an epidemic that’s spread across a very small demographical region? Will this help us, or will it cause problems?
In my opinion, centralizing, or converging all health data into a single processing system encourages the possibility of a single point of failure.
On the other hand, de centralizing will improve the system by offering it not one, but several chances to succeed. It will also let us create a tool that will support both pandemic and epidemic surveillance.
Another point – check out this [1] an article on ‘Rapid detection of pandemic influenza in the presence of seasonal influenza’
My question is, how do we identify if it’s just a seasonal ‘spike’ or an emerging epidemic? And how do we map symptoms to specific illnesses, and make that match before it’s too late?
So how to detect an emerging outbreak?
I came up with the following diagram to track patients and their symptoms for hospital A.
The three types of images given here depict different symptoms.
Assume the cases of Sam, Jack, Jim and Julian.
On Day 2, Sam, Jim and Julian all develop symptom A, which is associated with flu. So does this mean that we are facing an influenza epidemic? Maybe, maybe not. But on day three, Sam and Jim both develop symptom B. So now we have a pattern.
We also note that Sam and Jim go on to develop Symptom C on day 5. So this would definitely mean that Sam and Jim are suffering from the same illnesses.
But consider Jack. He developed symptom B a day later than normal. So what are his chances of suffering from the same illness as Sam and Jim?
And also, it seems that Julian has developed symptom A and C, but not B.
So based on this, and assuming that the symptoms are for flu, we could hypothetically come up with the following.
- Julian is suffering from ordinary flu
- Sam and Jim are suffering from Influenza.
- Jack is possibly suffering from Influenza.
So now we have a pattern, and we can stay alert for it. This pattern can also be used together with mathematical disease modeling to watch for outbreaks at other locations as well. This works well with the centralized (converged) vs. decentralized surveillance model I described earlier.
For example, assuming we have five data sources (hospitals) and one central surveillance unit.
If the unit fails to identify an epidemic incident in source A, it will essentially fail to identify similar cases of epidemics in the remaining four sources as well.
But if de centralized, we are improving the chances of epidemic detection by five times.
Example: assuming that our system will detect an epidemic in source A, but fail to do so in other four sources.
Once the pandemic is identified in source A, the unit will extract a set of core ‘definers’ that will be sent to the other units for further processing. Each of the other centers will do a backwards check to see if there is an indication of such a pattern in symptoms reported to them. Not only do these centers get a second chance to check for mistakes, they also get a pre warning if an outbreak is imminent.
And assuming that a disease has only just infected the region, a different location may have only received reports of symptom A so far. If so, due to the indicators sent from other regions they will be able to make preparations for a possible outbreak depending on the emergence of symptom B as well.
We may even share the core definers with other hospitals to do a wider ranging check.
[0] http://www.rand.org/pubs/research_briefs/RB9042/index1.html
[1] http://www.biomedcentral.com/content/pdf/1471-2458-10-726.pdf
Monday, September 19, 2011
My top OpenMRS videos
OpenMRS Co- founders Dr. Mamlin and Dr. Biondritch at Google tech talks. This video explains the original idea behind OpenMRS, and how the project actually got started. Its long, but very much worth watching.
This is a good place for all OpenMRS newbies to start. Its a very basic video that runs through how to set up OpenMRS environment and get started. I only wish I had paid it more attention to it at the time I was starting up for the first time :-)
Thursday, September 15, 2011
Welcome and Introduction
I plan to use this blog as a public diary on my path to improve my Health Informatics knowledge.
Health Informatics has been a long term passion of mine. Unfortunately, my home country does not have much to offer in this field, so I've decided to step out on my own, and to self educate myself to the best of my ability.
This blog is an attempt to record my 'Education', and will (hopefully) be useful to others who trod behind me.
My 'improvement' will be a community driven and long term process.
But be warned, Real work experaince will be my exams, and my friends in this industry will be my tutors
Where this journey will take me, and how far I can achieve my goals, only time will tell.
Subscribe to:
Posts (Atom)
